Research projects
Our research interest is to elucidate the biological significance of networks emerging out of living organisms. We, as living organisms, have contradictory features, universality and uniquness, which are ultimately controlled by genes encoded in a genome. Thus, we had been thinking that understanding functions of genes resolved mechanisms how living organisms orchestrate universality and uniquness. However, our system is more complicated. Genes are indeed fundamental, but tremendous amounts of combination of the genes are involved in our system. Such a combination, "Network", is central to the orchestration of biological system. Therfore, we are interested in elucidation of network emerging out of living organisms. In particular, we are currently focusing on Gene Regulatory Network (GRN) involved in development, metablisms and ineter-kingdom associations.
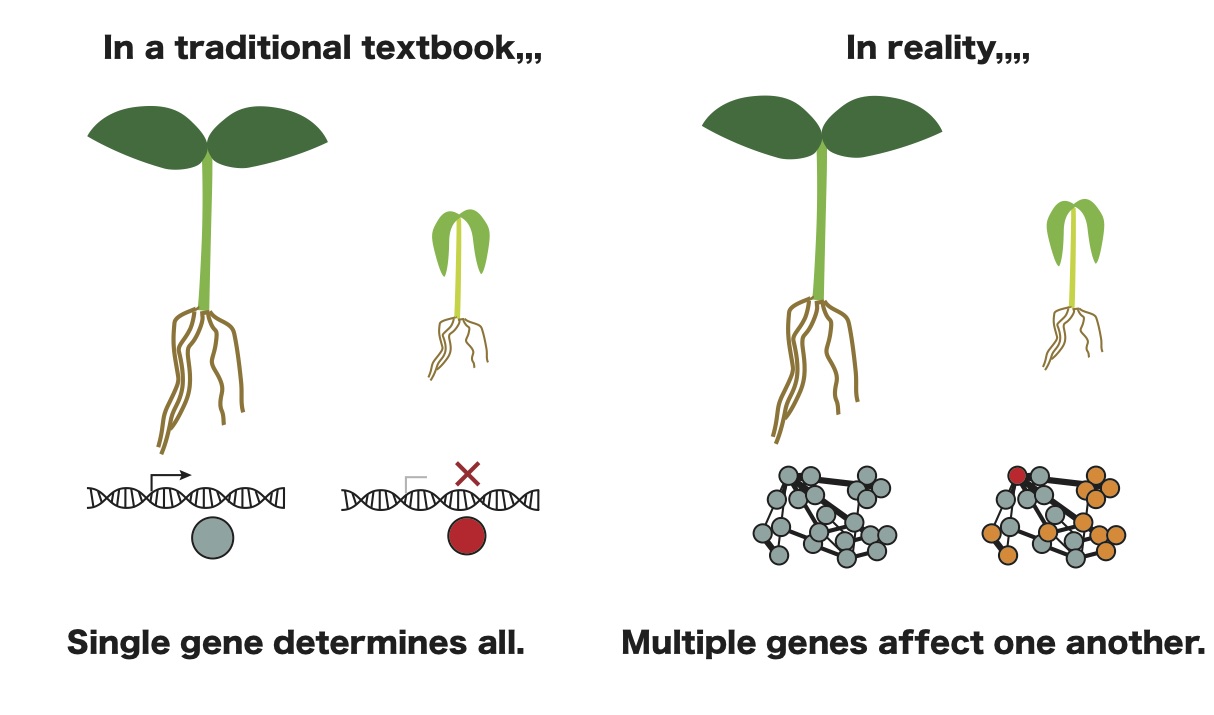
1. GRNs involving flavonoids on epidermal cell development
Elucidation of biological processes through the analysis of varied molecular networks has proven highly valuable with scrutiny at the DNA, RNA, protein, and metabolite levels each providing distinct and complementary information. In our future research, we will use our expertise in biological networks for agricultural benefit, in particular by investigating gene regulatory network associated with secondary metabolites.
Plants produce secondary metabolites that significantly affect biological systems in diverse organisms. More than 200,000 secondary metabolites have been reported to date, consisting of various types of compounds, such as terpenoids, alkaloids, and phenolics (Verpoorte and Memelink, 2002; Zwenger and Basu, 2013). Flavonoids, a type of phenolic, function affect various biological processes in both plants and animals (Havsteen, 2002; Taylor and Grotewold, 2005). In plants, flavonoids play as signaling molecules in development and plant-microbe interactions (Taylor and Grotewold, 2005).
Notably, flavonoids alter gene expression patterns (Ringli et al., 2008; Rohde et al., 2004). Plants with a mutation in an enzyme involved in the flavonoid pathway change both flavonoid profiles and gene expression patterns in a genome-wide manner (Pourcel et al., 2013; Ringli et al., 2008; Rohde et al., 2004). Indeed, the flavonoids are located in the nucleus (Saslowsky, 2005), suggesting that flavonoids directly regulate gene expression via interactions with transcription factors (TFs). Flavonoids are known to associate with a broad range of proteins (Arango, Morohashi et al., 2013; Gledhill et al., 2007), increasing the likelihood that direct flavonoid-TF binding occurs.
Despite the importance of flavonoids in development as well as plant-microbe interaction, the roles of flavonoids in the regulation of gene expression by gene regulatory networks (GRNs) is largely unknown. Our research projects will focus on the role of flavonoid-GRN relationships in plant development and plant-microbe interactions. To elucidate the effects of flavonoids on the GRNs, we will take an advantage of systems biology approaches that consist of three main projects according to the flavonoid functions in A) development, B) plant-microbe interaction, and C) direct perturbation of gene expression as detail below
To address the question of how flavonoids affect plant development via changes in gene expression, our research will focus on two organs that develop from epidermal cells: trichomes and root hair cells in Arabidopsis thaliana. Genes involved in the development of trichomes and root hairs in Arabidopsis thaliana significantly overlap (Ishida et al., 2008).
Among the genes involved in both developmental events, GLABRA3 (GL3) encodes a pivotal, basic helix-loop-helix (bHLH) TF (Zhang et al., 2003). Our previous studies, as well as several others, have revealed the GRNs involving GL3 in the development of trichomes and root hair cells (Bruex et al., 2012; Morohashi and Grotewold, 2009; Morohashi et al., 2007). Interestingly, GL3 also plays an indispensable role in flavonoid pathways (Gonzalez et al., 2008). While trichomes and root hair cells accumulate flavonoids (Hassan and Mathesius, 2012; Schilmiller et al., 2010; Sinlapadech et al., 2007), a connection between flavonoid production and GRNs is unknown. Our current research strongly suggests that the GRN involving GL3 integrates both developmental and flavonoid pathway via small RNAs. I will expand the current research to explore the role of flavonoids in the GRNs underlying the epidermal cell development.
2. Comprehensively identifying flavonoid-associated transcription factors in plants
Flavonoid-associated TFs likely play pivotal roles in development as well as PGPR-plant interactions. However, identification of flavonoid-associated TFs is technically challenging since affinities between small molecules and proteins are relatively low. To overcome the problem, we will use an innovative method called phage display coupled with next-generation sequencing (PD-Seq), which I have developed to successfully dissect individual metabolite- protein interactions (Arango, Morohashi et al., 2013). Successful identification of flavonoid-associated TFs will allow us to build a directional GRN by applying ChIP-Seq, also within our expertise, further elucidating the biological processes underlying development and PGPR-plant interactions.
3. GRNs underlying flavonoid-dependent PGPR endophytic colonizations
Flavonoids increase the growth of a certain type of plant growth-promoting bacterium (PGPR), which benefits plant growth via endophytic associations with a broad range of plant species such as maize, rice, sorghum, sugarcane, and Arabidopsis thaliana (Gough et al., 1997; Monteiro et al., 2012). We are using Arabidopsis thaliana and Zea mays as dicot and monocot models, respectively, to investigate the flavonoid-dependent PGPR interaction. Network analysis will determine the effects of PGPR on host plant gene expression and gene expression in PGPR in the presence and absence of flavonoids. By precisely observing the changes in gene expression over the course of root colonization, we will reveal the dynamics of the flavonoid- induced GRN. Since I have revealed the GRNs of the flavonoid pathway in A. thaliana and Z. mays by a systems approach (Morohashi et al., 2012; Morohashi and Grotewold, 2009), we have the ability to uncover the GRNs in plant-microbe interaction as well.
References
- Arango, D*., Morohashi*, K., Yilmaz, A., Kuramochi, K., Parihar, A., Brahimaj, B., Grotewold, E., and Doseff, A.I. (2013). Molecular basis for the action of a dietary flavonoid revealed by the comprehensive identification of apigenin human targets. Proc Natl Acad Sci USA. 110, E2153–E2162.
- Bruex, A., Kainkaryam, R.M., Wieckowski, Y., Kang, Y.H., Bernhardt, C., Xia, Y., Zheng, X., Wang, J.Y., Lee, M.M., Benfey, P., et al. (2012). A gene regulatory network for root epidermis cell differentiation in Arabidopsis. PLoS Genet 8, e1002446.
- Gledhill, J.R., Montgomery, M.G., Leslie, A.G.W., and Walker, J.E. (2007). Mechanism of inhibition of bovine F1- ATPase by resveratrol and related polyphenols. Proc Natl Acad Sci USA. 104, 13632–13637.
- Gonzalez, A., Zhao, M., Leavitt, J.M., and Lloyd, A.M. (2008). Regulation of the anthocyanin biosynthetic pathway by the TTG1/bHLH/Myb transcriptional complex in Arabidopsis seedlings. Plant J 53, 814–827.
- Gough, C., Galera, C., Vasse, J., Webster, G., Cocking, E.C., and Denarie, J. (1997). Specific flavonoids promote intercellular root colonization of Arabidopsis thaliana by Azorhizobium caulinodans ORS571. Mol Plant Microbe Interact 10, 560–570.
- Hassan, S., and Mathesius, U. (2012). The role of flavonoids in root-rhizosphere signalling: opportunities and challenges for improving plant-microbe interactions. J Exp Bot 63, 3429–3444.
- Havsteen, B.H. (2002). The biochemistry and medical significance of the flavonoids. Pharmacol Ther 96, 67–202. Ishida, T., Kurata, T., Okada, K., and Wada, T. (2008). A genetic regulatory network in the development of trichomes and root hairs. Ann Rev Plant Biol 59, 365–386.
- Monteiro, R.A., Balsanelli, E., Wassem, R., Marin, A.M., Brusamarello-Santos, L.C.C., Schmidt, M.A., Tadra-Sfeir, M.Z., Pankievicz, V.C.S., Cruz, L.M., Chubatsu, L.S., et al. (2012). Herbaspirillum-plant interactions: microscopical, histological and molecular aspects. Plant and Soil 356, 175–196.
- Morohashi, K., Casas, M.I., Falcone Ferreyra, M.L., Mejia-Guerra, M.K., Pourcel, L., Yilmaz, A., Feller, A., Carvalho, B., Emiliani, J., Rodriguez, E., et al. (2012). A genome-wide regulatory framework identifies maize Pericarp Color1 controlled genes. Plant Cell 24, 2745–2764.
- Morohashi, K., and Grotewold, E. (2009). A systems approach reveals regulatory circuitry for Arabidopsis trichome initiation by the GL3 and GL1 selectors. PLoS Genet 5, e1000396.
- Morohashi, K., Zhao, M., Yang, M., Read, B., Lloyd, A., Lamb, R., and Grotewold, E. (2007). Participation of the Arabidopsis bHLH factor GL3 in trichome initiation regulatory events. Plant Physiol. 145, 736–746.
- Pourcel, L., Irani, N.G., Koo, A.J.K., Bohorquez-Restrepo, A., Howe, G.A., and Grotewold, E. (2013). A chemical complementation approach reveals genes and interactions of flavonoids with other pathways. Plant J 74, 383–397.
- Ringli, C., Bigler, L., Kuhn, B.M., Leiber, R.-M., Diet, A., Santelia, D., Frey, B., Pollmann, S., and Klein, M. (2008). The modified flavonol glycosylation profile in the Arabidopsis rol1 mutants results in alterations in plant growth and cell shape formation. Plant Cell 20, 1470–1481.
- Rohde, A., Morreel, K., Ralph, J., Goeminne, G., Hostyn, V., De Rycke, R., Kushnir, S., Van Doorsselaere, J., Joseleau, J.-P., Vuylsteke, M., et al. (2004). Molecular phenotyping of the pal1 and pal2 mutants of Arabidopsis thaliana reveals far-reaching consequences on phenylpropanoid, amino acid, and carbohydrate metabolism. Plant Cell 16, 2749–2771.
- Saslowsky, D.E. (2005). Nuclear localization of flavonoid enzymes in Arabidopsis. J Biol Chem 280, 23735–23740. Schilmiller, A.L., Miner, D.P., Larson, M., McDowell, E., Gang, D.R., Wilkerson, C., and Last, R.L. (2010). Studies of a biochemical factory: tomato trichome deep expressed sequence tag sequencing and proteomics. Plant Physiol 153, 1212–1223.
- Sinlapadech, T., Stout, J., Ruegger, M.O., Deak, M., and Chapple, C. (2007). The hyper-fluorescent trichome phenotype of the brt1 mutant of Arabidopsis is the result of a defect in a sinapic acid: UDPG glucosyltransferase. Plant J 49, 655–668.
- Taylor, L.P., and Grotewold, E. (2005). Flavonoids as developmental regulators. Curr Opin Plant Biol 8, 317–323.
- Verpoorte, R., and Memelink, J. (2002). Engineering secondary metabolite production in plants. Curr Opin Biotechnol 13, 181–187.
- Zhang, F., Gonzalez, A., Zhao, M., Payne, C.T., and Lloyd, A. (2003). A network of redundant bHLH proteins functions in all TTG1-dependent pathways of Arabidopsis. Development 130, 4859–4869.
- Zwenger, S., and Basu, C. (2013). Plant terpenoids: applications and future potentials. Biotechnol Mol Biol Rev 3, 1–7.

